Elena Shagimardanova, MSc, PhD
Ruslan Devyatiyarov, MSc
Marat Sabirov, Ph.
Anastasia Gainullina, PhD
Vasily Sokolov, MSc
Chikina Evgenia, MSc
Zakhar Starrinov, MSc
The ability to adapt to unfavorable environmental conditions has led to the emergence in a number of organisms of unique adaptations at the molecular genetic level that provide effective defense in extreme conditions. Using modern genetic technologies, we explore examples of such adaptations, characterizing key biomolecules with useful properties for use in biomedicine and agriculture.
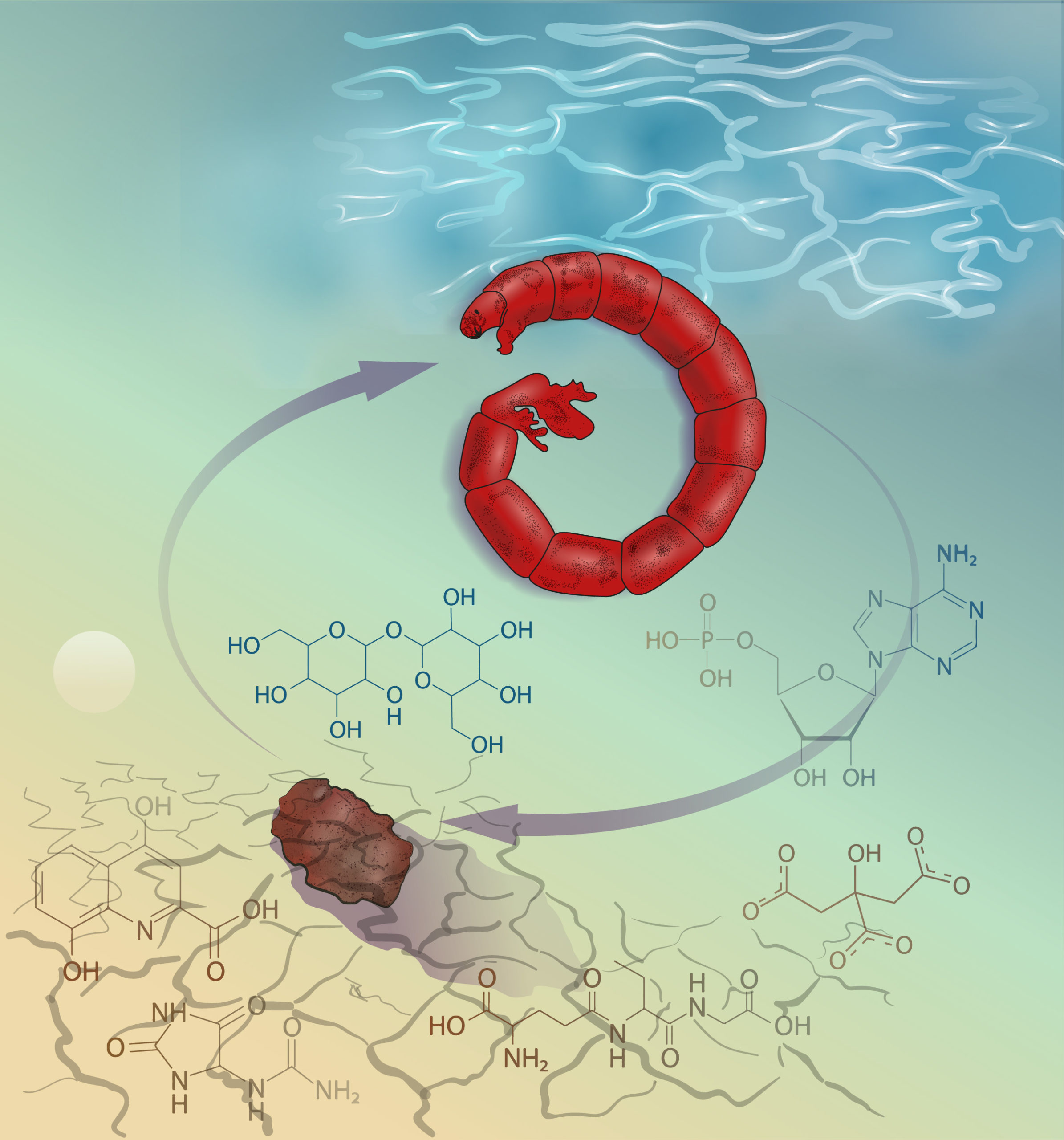
A number of organisms are capable of completely shutting down metabolism while effectively protecting cells and tissues, including the brain, from damage while surviving unfavorable periods of extreme environmental change. We investigate two examples of such adaptations: anhydrobiosis of Polypedilum vanderplanki mosquito larvae withstanding complete dehydration and cryobiosis of a number of invertebrate organisms withstanding instantaneous deep freezing. Modern methods of comparative whole-genome analysis, in combination with the study of RNA transcription in individual cells, allow us to highlight key evolutionary events that led to the emergence of effective defenses in the cells of such organisms. Our goal is to use knowledge of the mechanisms of anhydrobiosis and cryobiosis to develop technologies for anhydrous storage of living cells and tissues, as well as methods to reduce the negative effects of reperfusion syndrome and other post-operative complications in humans.
The ability to regenerate organs and tissues is limited in humans and most mammals. At the same time, needle-nosed mice of the genus Acomys restore the loss of more than 60% of skin cover after traumatization and are distinguished by unusual abilities to regenerate skeletal muscles, spinal cord, cartilaginous tissue and other organs. In addition, this group of animals are characterized by complex social behavior, which is associated with the structure of the sympathetic nervous system, including the adrenal glands, which are similar in structure to human. Using methods of comparative evolutionary genomics, gene transcription analysis including scRNASeq, proteomic and metabolomic analysis we are studying the origin and molecular mechanisms of regeneration in needle-nosed mice, with the goal of using this knowledge for biomimetic technologies to improve wound healing, stimulate tissue repair processes and enhance human regenerative potential in the future.
Periods of temporary hypometabolism such as hibernation and embryonic diapause are of great interest in terms of molecular mechanisms of stress response regulation and the study of genetic control of cellular and individual organelle activity. We use a number of models of hypometabolism, including the torpor of avian embryos to understand the molecular genetic mechanisms of temporary hypometabolism and the reversible control of organismal dynamics. Based on multi-omics analysis of the processes occurring in animals in the state of hypometabolism, we are developing approaches to initiate artificial hibernation for biomedical purposes in the future.
Protein-coding regions account for only a few percent of the genome. At the same time, noncoding regulatory regions play a key role in the regulation of genetic networks. Non-coding RNAs associated with enhancer and promoter regions attract more and more attention of researchers, as more than 80% of mutations associated with human diseases are located in these regions. Using original methods of RNA analysis (including at the level of individual cells) and high-throughput sequencing, we create digital genetic resources and study the influence of regulatory regions of the genome on the pathogenesis of various diseases, gene function in various tissues and various phenotypic traits of humans and animals. Our research is related to the use of genome regulatory regions in animal breeding, medical genetics, sports and extreme medicine.
Main LIFT-affiliated publications:
Population Genomics of Two Closely Related Anhydrobiotic Midges Reveals Differences in Adaptation to Extreme Desiccation, Shaikhutdinov N., Klink G., Garushyants S., Kozlova O., Cherkasov A., Kikawada T., Okuda T., Pemba D., Shagimardanova E., Penin A., Deviatiiarov R., Gazizova G., Cornette R., Gusev O., Bazykin G.. Genome Biology and Evolution, 15,10, 2023, evad169, https://doi.org/10.1093/gbe/evad169
Dosage Compensation of Z Sex Chromosome Genes in Avian Fibroblast Cells. Deviatiiarov, R., Nagai, H., Ismagulov, G. et al. Genome Biology 2023, 24, 213. https://doi.org/10.1186/s13059-023-03055-z
Biomimetic Cardiac Tissue Models for In Vitro Arrhythmia Studies. Aitova, A.; Berezhnoy, A.; Tsvelaya, V.; Gusev, O.; Lyundup, A.; Efimov, A.E.; Agapov, I.; Agladze, K. Biomimetics 2023, 8, 487. https://doi.org/10.3390/biomimetics8060487
Other publications:
A Bird’s-Eye View of Endangered Species Conservation: Avian Genomics and Stem Cell Approaches for Green Peafowl (Pavo muticus). Intarapat, S.; Sukparangsi, W.; Gusev, O.; Sheng, G. Genes 2023, 14, 2040. https://doi.org/10.3390/genes14112040
LIFT Center LLC
Address: 121205, Moscow, territory of the Skolkovo Innovation Center, Skolkovo, Moscow.
Skolkovo Innovation Center,
ul. 5 Nobel str.